Biography
Michelle Ward received her M.Sc. from the University of Cape Town where she conducted research aimed at identifying a novel therapeutic target in cervical cancer, the second most common form of cancer in women in South Africa. After studying the regulation of a single gene, she became interested in understanding the general ‘rules’ governing the regulation of transcription genome-wide. For her Ph.D., she therefore joined Dr. Duncan Odom’s group at the University of Cambridge as a Commonwealth Scholar to learn genomic approaches for investigating global gene regulation. These methodologies allowed her to gain insight into the evolution of CTCF binding sites across multiple primate species, and her Ph.D. thesis focused on investigating the regulatory potential of transposable elements (TEs) in mammalian genomes. This work suggests that TEs contain intrinsic regulatory potential that can be revealed in specific contexts. To investigate the context-dependent regulation of the genome using a flexible induced pluripotent stem cell-based system, she joined Prof. Yoav Gilad’s group at the University of Chicago as an EMBO Long-Term Postdoctoral Fellow. Here she continued to investigate the regulation of TEs across species in pluripotent stem cells before establishing an in vitro system to study the effects of oxygen deprivation on stem cell-derived cardiomyocytes from humans and chimpanzees. She was recruited to UTMB as an Assistant Professor and CPRIT Scholar in Cancer Research in 2020.
Research
The goal of the Ward lab is to dissect the global role of regulatory elements, including those derived from TEs, in directing gene expression in healthy, stressed and disease states in cardiovascular disease-relevant cell types. We use a variety of functional genomics, and induced pluripotent stem cell technology-based approaches to tackle this problem.
The three areas we are currently investigating are:
1) Gene regulatory dynamics during differentiation to cardiovascular cell types
Global gene regulatory networks and mechanisms, and their evolutionary robustness, are not fully understood in complex processes such as development, especially in humans and other primates. In vitro cellular differentiation assays across species allow for the study of gene regulatory cascades from induced pluripotent stem cells (iPSCs), to intermediate germ layers, and terminal cell types. The regulation of, and by, TEs in this process is of particular interest to us. This research area will lead to insight into the gene regulatory dynamics involved in the transitions between cell states during differentiation.
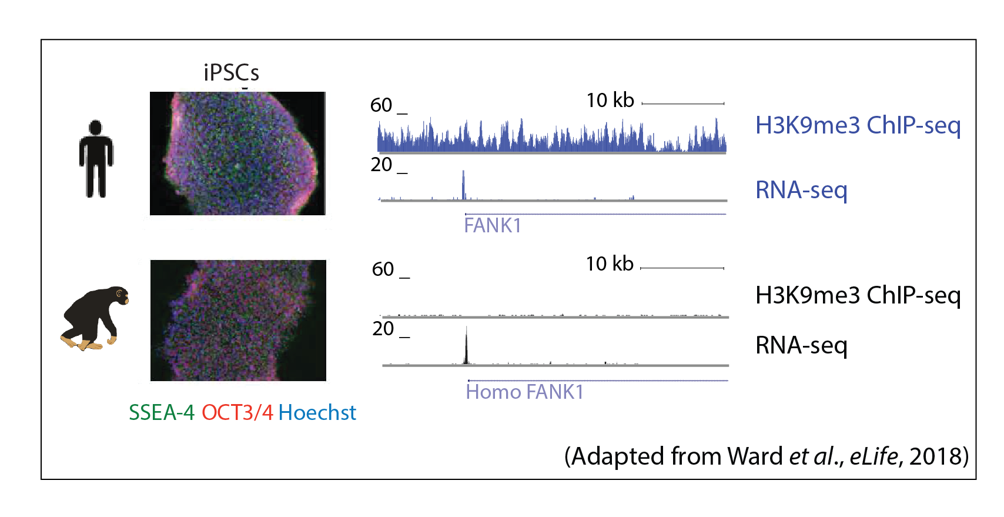
Relevant publications:
Ward, M.C.#, Zhao, S., Luo, K., Pavlovic, B.J., Karimi, M.M., Stephens, M., Gilad, Y#. Silencing of transposable elements may not be a major driver of regulatory evolution in primate induced pluripotent stem cells. eLife (2018) 10.7554/eLife.33084.
Ward, M.C.*, Wilson, M.D.*, Barbosa-Morais, N.L., Schmidt, D., Stark, R., Pan, Q., Schwalie, P.C., Menon, S., Lukk, M., Watt, S., Thybert, D., Kutter, C., Kirschner K., Flicek, P., Blencowe, B.J. and Odom, D.T. Latent regulatory potential of human-specific repetitive elements. Molecular Cell (2013) 262-272.
Schwalie, P.C.*, Ward, M.C.*, Cain, C.E., Faure, A.J., Gilad, Y., Odom, D.T. and Flicek, P. Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primates. Genome Biology (2013) 14:R148.
2) Robustness of gene regulatory processes in response to perturbation
Rapid changes in gene expression in response to stress can lead to evolutionary adaptation. If these changes occur in a species-specific manner, they could have consequences on inter-species differences in cellular phenotypes. Alternatively, there may be redundancy in gene regulatory networks. This notion has been difficult to investigate between primate species due to practical and ethical challenges. In addition, teasing apart the relative contributions of intrinsic genetic sequence and the environmental context is inherently challenging in identifying the source of any inter-species differences in gene expression. iPSCs, equivalently generated across species, now allow for environmentally controlled studies to determine core inter-species gene regulatory differences across multiple cell types. This research area will lead to insight into the robustness of gene regulatory networks across cell types, and under different environmental conditions and stress.
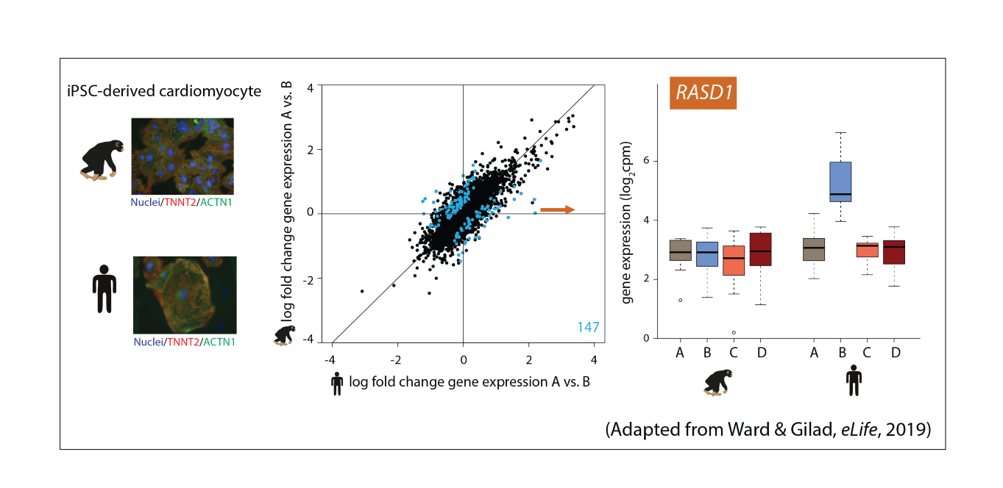
Figure 2: Assaying the response to stress in iPSC-derived cardiomyocytes from humans and chimpanzees.
Relevant publications:
Ward, M.C.# and Gilad, Y.# A generally conserved response to hypoxia in iPSC-derived cardiomyocytes from humans and chimpanzees. eLife (2019) https://doi.org/10.7554/eLife.42374.
3) Impact of inter-individual variation on cardiovascular disease-relevant phenotypes
Genetic variants have been found to be associated with a variety of phenotypic traits and diseases, including cardiovascular disease. These loci are typically in non-coding regions of the genome. However, many of these variants are also eQTLs, which could implicate particular genes involved in the trait. Understanding the impact of genetic variation in the human population will provide insight into disease susceptibility and response to treatment, central tenets of personalized medicine. Characterizing the influence of genetic variants, in the relevant cell type, in a controlled environmental context will be important in working towards this goal. One particular area of interest focuses on understanding the genetic and mechanistic basis of chemotherapeutic agent-induced cardiotoxicity. This research area will lead to insight into the effects of genetic variation on drug responses and CVD phenotypes, and could help inform appropriate patient treatment.
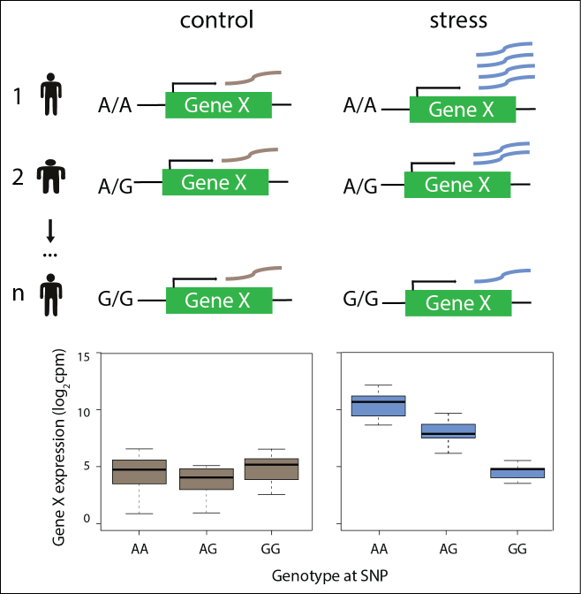
Figure 3: Assaying the effects of genetic variation on gene expression following stress in human cardiomyocytes.
Relevant publications:
Ward, M.C.*#, Banovich, N.E.*, Sarkar, A., Stephens, M., Gilad, Y#. Dynamic effects of genetic variation on gene expression revealed following hypoxic stress in cardiomyocytes. biorxiv (2020) https://doi.org/10.1101/2020.03.28.012823
Banovich, N.E.*, Li, Y.I.*, Raj, A.*, Ward, M.C., Greenside, P., Calderon, D., Tung, P.Y., Burnett, J.E., Myrthil, M., Thomas, S.M., Burrows, C.K., Gallego Romero, I., Pavlovic, B.J., Kundaje, A., Pritchard, J.K., Gilad, Y. Impact of regulatory variation across human iPSCs and differentiated cells. Genome Research (2018) 122-131.
If you are interested in working with us in one of these three areas please contact Michelle Ward by email for details on current open positions.