Research Areas
Biofilms, Host-pathogen interactions, Salmonella lifestyle switch, Salmonella carriage and dormancy
Research Interests
A fundamental trait shared amongst many mammalian pathogens is their ability to exist in different forms in order to survive and replicate in often fluctuating host and non-host environments. Two important gut pathogens, Salmonella Typhimurium and Salmonella Typhi, which cause gastroenteritis and typhoid fever, respectively, survive inside host cells by activation of a specific class of virulence genes. Salmonellae also develop to form hardy multi-cellular communities, called biofilms, on host tissues for successful survival and persistence in asymptomatic carriers. An active area of research focuses on understanding how the genetic program is regulated in coordination with environmental changes to evoke lifestyle changes in vivo. 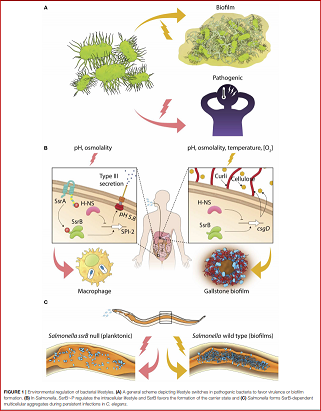
We discovered that SsrB, the transcriptional regulator of virulence genes, has the additional function of inducing biofilm formation by activating expression of the central biofilm regulator, CsgD (Desai et al., eLife, 2016). Generally, specific environmental changes activate a membrane-bound kinase, which in turn transfers the phosphoryl group to a transcriptional regulator in order to regulate virulence gene transcription. But we discovered that even without phosphorylation, SsrB retains the ability to bind DNA and relieve the H-NS-mediated repressive effect on csgD transcription. SsrB is thus a molecular switch which drives the twin lifestyles of Salmonella Typhimurium; as SsrB~P, when phosphorylated by an activated SsrA kinase, it activates around 30 virulence genes required for survival within the host; and as SsrB, in the case when SsrA is inactive or absent, it counters transcriptional silencing at csgD to form biofilms.
Interestingly, such a SsrB-driven lifestyle switch was also responsible for establishing dormancy in the intestines of persistently infected Caenorhabditis elegans (Desai et al., PNAS, 2019). We found that formation of biofilms in the intestinal lumen, dampened virulence and delayed worm death by activation of the mitogen-activated protein kinase (MAPK) innate immunity pathway. Overall, our understanding of how Salmonella establishes dormancy in persistent infections has been greatly enabled by the use of C. elegans as a model host, since the murine model is not conducive for real-time high-resolution microscopy. Future investigations will involve studying the mechanisms of lifestyle changes in more aggressive strains of Salmonella and deciphering the underlying signaling pathways using heterologous hosts as model systems.
Image credit: Melanie Lee, Science Communications Core, Mechanobiology Institute