Research Areas
Super-resolution imaging in bacteria and single cell analysis; Use of heterologous host models for Salmonella pathogenesis, including C. elegans and Zebrafish; Developing the oncolytic potential of Salmonella; Salmonella Typhi pathogenesis; Signal transduction in bacteria; Mechanotransduction and osmotic signaling in E. coli
Research Interests
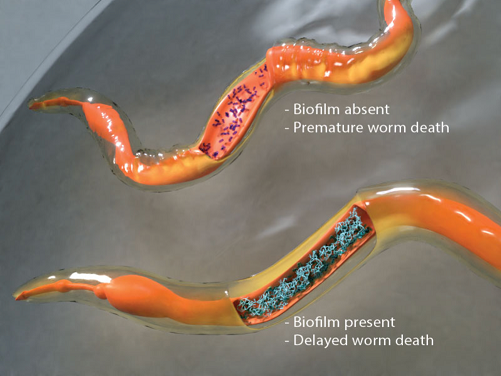
Salmonella can adopt either a virulent or a dormant lifestyle inside its host. What makes Salmonella such an expert pathogen is its ability to switch lifestyles depending on the environmental conditions that it encounters. Under favorable conditions (e.g. the acid pH of the vacuole), Salmonella activates its repertoire of virulence genes, which allows it to invade host cells and actively spread infection (Liew et al., eLife 2019). Under neutral conditions, dormancy genes are activated that allow the bacteria to lay low and form clusters called biofilms. The two-component response regulator SsrB acts canonically through phosphorylation from its kinase SsrA to activate virulence genes, and non-canonically when the SsrA kinase is at very low levels to de-repress H-NS and drive activation of the biofilm master regulator CsgD (Desai et al. eLife, 2016), reviewed in (Desai & Kenney, 2017, 2019). Dormant states have been characterized by the formation of biofilms inside the host and the molecular mechanisms that lead to the formation of biofilm inside live worm hosts were identified in our recent study (Desai et al., PNAS 2019) (see Figure). This work uncovers how the molecular mechanisms that drive biofilm formation under laboratory conditions are equally relevant in living systems and it now allows us to translate laboratory knowledge into tools for tackling bacterial persistence and pathogenesis in living hosts.
Our most recent project involves developing the oncolytic potential of Salmonella. It is capable of targeting tumors and causing tumor regression. In order to understand the fundamentals of tumor targeting and regression, we are combining spheroids, organoids and heterologous host models, including: C. elegans, Zebrafish and the mouse Lastly, our laboratory has a long-standing interest in signal transduction and the regulation of gene expression in prokaryotes. In particular, we study the two-component regulatory system EnvZ/OmpR that regulates the expression of outer membrane proteins, as well as many other genes involved in acid and osmotic stress. Our present work focuses on how OmpR activates genes required for systemic infection located on Salmonella pathogenicity island 2 in Salmonella enterica.
Biography
Dr Kenney was a Professor of Microbiology at the University of Illinois-Chicago and a Visiting Professor at the Mechanobiology Institute, National University of Singapore until 2019, when she was recruited to UTMB. Her laboratory studies Salmonella pathogenesis, its application as an oncolytic agent and two-component signaling systems in bacteria that control gene expression at single cell and nanometer levels.
Recent Publications
- Desai, S.K. and L.J. Kenney. Switching lifestyles is an in vivo adaptation strategy of bacterial pathogens. Front. Cell. Infect. Microbiol. 9:421. doi: 10.3389/fcimb.2019.00421
- Kenney, L.J. and G.S. Anand. EnvZ/OmpR two-component signaling: an archetype system that can function non-canonically. EcoSalPlus. 2020.
- Singh, M.K. and L.J. Kenney. Super-resolution imaging of bacterial pathogens and visualization of their secreted effectors. FEMS Microbiol. Rev. In Press. 2020.
- Desai SK, Padmanabhan A, Harshe S, Zaidel-Bar R, and Kenney LJ. Salmonella biofilms program innate immunity for persistence in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 2019;. [PMID: 31160462]
- Ottemann KM, and Kenney LJ. Editorial overview: Host-pathogen interactions: bacteria. Curr. Opin. Microbiol. 2019; 47:iii-v. [PMID: 31138403]
- Liew ATF, Foo YH, Gao Y, Zangoui P, Singh MK, Gulvady R, and Kenney LJ. Single cell, super-resolution imaging reveals an acid pH-dependent conformational switch in SsrB regulates SPI-2. Elife 2019; 8. [PMID: 31033442]
- Chakraborty S, and Kenney LJ. A New Role of OmpR in Acid and Osmotic Stress in Salmonella and E. coli. Front Microbiol 2018; 9:2656. [PMID: 30524381]
- Kenney LJ. The role of acid stress in Salmonella pathogenesis. Curr. Opin. Microbiol. 2018; 47:45-51. [PMID: 30529007]
- Ghosh M, Wang LC, Huber R, Gao Y, Morgan LK, Tulsian NK, Bond P, Kenney LJ, and Anand GS. Engineering an Osmosensor by Pivotal Histidine Positioning within Disordered Helices. Structure 2018;. [PMID: 30503779]
- Gulvady R, Gao Y, Kenney LJ, and Yan J. A single molecule analysis of H-NS uncouples DNA binding affinity from DNA specificity. Nucleic Acids Res. 2018;. [PMID: 30239908]
- Mather AE, Phuong TLT, Gao Y, Clare S, Mukhopadhyay S, Goulding DA, Hoang NTD, Tuyen HT, Lan NPH, Thompson CN, Trang NHT, Carrique-Mas J, Tue NT, Campbell JI, Rabaa MA, Thanh DP, Harcourt K, Hoa NT, Trung NV, Schultsz C, Perron GG, Coia JE, Brown DJ, Okoro C, Parkhill J, Thomson NR, Chau NVV, Thwaites GE, Maskell DJ, Dougan G, Kenney LJ, and Baker S. New Variant of Multidrug-Resistant Salmonella enterica Serovar Typhimurium Associated with Invasive Disease in Immunocompromised Patients in Vietnam. MBio 2018; 9(5). [PMID: 30181247]
- Gao Y, Spahn C, Heilemann M, and Kenney LJ. The Pearling Transition Provides Evidence of Force-Driven Endosomal Tubulation during Salmonella Infection. MBio 2018; 9(3). [PMID: 29921673]
- Dykas MM, Desai SK, Patra A, Motapothula MR, Poddar K, Kenney LJ, and Venkatesan T. Identification of Biofilm Inhibitors by Screening Combinatorial Libraries of Metal Oxide Thin Films. ACS Appl Mater Interfaces 2018;. [PMID: 29553712]